📑 Learning Objectives
- Applying the Kernel Ridge Regression model with
scikit-learn - Exploring hyperparameters and kernel choices
- Visualising output
Linear models are too coarse for subtle data trends
We've chewed over the theory. Now let's see how this technique may be applied to a visual regression problem. Let's begin by generating some data.
RANDOM_SEED = 31
NOISE = 2
SCALE = 20
# Define the ground truth function
x = np.linspace(0, SCALE, 101)
y_true = 1.5 * (x -7 + 2*np.sin(x) + np.sin(2*x))
# Generate data skewed with noise
ep = NOISE * np.random.normal(size=(len(x),))
y = y_true + ep
# Columns
x = x[:, np.newaxis]
y = y[:, np.newaxis]
Here we consider the data-generating (ground truth) function
which is skewed, as , with some normally distributed noise . To see this data, let's fit a linear regression through the data points and plot the results.
from sklearn.linear_model import LinearRegression
linear_model = LinearRegression()
linear_model.fit(x, y)
print('Score: ', linear_model.score(x, y))
print('Coeff: ', linear_model.coef_)
print('Intercept: ', linear_model.intercept_)
x_test = np.linspace(0, SCALE, 1001)
x_test = x_test[:, np.newaxis]
y_pred = linear_model.predict(x_test)
y_gt = 1.5 * (x_test -7 + 2*np.sin(x_test) + np.sin(2*x_test))
scat = plt.scatter(x, y, marker='.')
line, = plt.plot(x_test, y_pred, 'g', linewidth=2)
true, = plt.plot(x_test, y_gt, '--r')
plt.legend([scat, line, true], ['Data', 'Prediction', 'Ground Truth'])
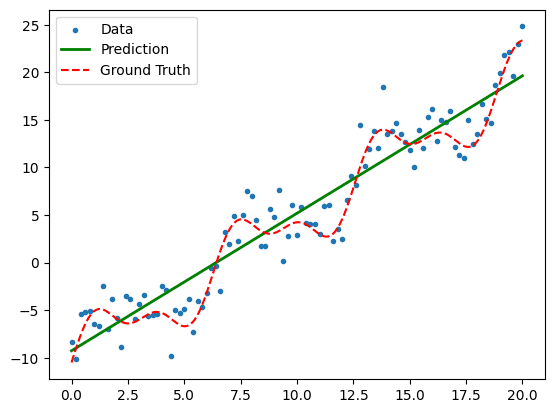
The linear fit produces a coefficient of determination (the R^2 value) of 0.90.
Not bad, but given the non-linear form of the ground truth function, we have a strong
prior belief that the model has not captured all of the features.
Kernel regression can identify more features by high-dimensional projections
Now let us try a variety of kernel models to detect these features.
from sklearn.kernel_ridge import KernelRidge
# Polynomial kernel
krr_poly_model = KernelRidge(kernel='polynomial', degree=5)
krr_poly_model.fit(x, y)
# Radial Basis Function kernel
krr_rbf_model = KernelRidge(kernel='rbf')
krr_rbf_model.fit(x, y)
# Laplacian kernel
krr_laplacian_model = KernelRidge(kernel='laplacian', gamma=1/SCALE)
krr_laplacian_model.fit(x, y)
# Linear kernel
krr_linear_model = KernelRidge(kernel='linear')
krr_linear_model.fit(x, y)
# Test points
x_test = np.linspace(0, SCALE, 1001)
x_test = x_test[:, np.newaxis]
# Predictions
y_poly = krr_poly_model.predict(x_test)
y_rbf = krr_rbf_model.predict(x_test)
y_laplacian = krr_laplacian_model.predict(x_test)
y_linear = krr_linear_model.predict(x_test)
# Ground truth
y_gt = 1.5 * (x_test -7 + 2*np.sin(x_test) + np.sin(2*x_test))
# Plot results
fx, ax = plt.subplots(2, 2, figsize=(10, 10))
# Linear kernel
scat = ax[0, 0].scatter(x, y, marker='.')
line, = ax[0, 0].plot(x_test, y_linear, 'g', linewidth=2)
true, = ax[0, 0].plot(x_test, y_gt, '--r')
ax[0, 0].legend([scat, line, true], ['Data', 'Prediction', 'Ground Truth'])
ax[0, 0].set_title('Linear Kernel')
# Polynomial kernel
scat = ax[0, 1].scatter(x, y, marker='.')
line, = ax[0, 1].plot(x_test, y_poly, 'g', linewidth=2)
true, = ax[0, 1].plot(x_test, y_gt, '--r')
ax[0, 1].legend([scat, line, true], ['Data', 'Prediction', 'Ground Truth'])
ax[0, 1].set_title('Degree 5 Polynomial Kernel')
# Laplacian kernel
scat = ax[1, 0].scatter(x, y, marker='.')
line, = ax[1, 0].plot(x_test, y_laplacian, 'g', linewidth=2)
true, = ax[1, 0].plot(x_test, y_gt, '--r')
ax[1, 0].legend([scat, line, true], ['Data', 'Prediction', 'Ground Truth'])
ax[1, 0].set_title('Laplacian Kernel')
# RBF kernel
scat = ax[1, 1].scatter(x, y, marker='.')
line, = ax[1, 1].plot(x_test, y_rbf, 'g', linewidth=2)
true, = ax[1, 1].plot(x_test, y_gt, '--r')
ax[1, 1].legend([scat, line, true], ['Data', 'Prediction', 'Ground Truth'])
ax[1, 1].set_title('Radial Basis Function Kernel')
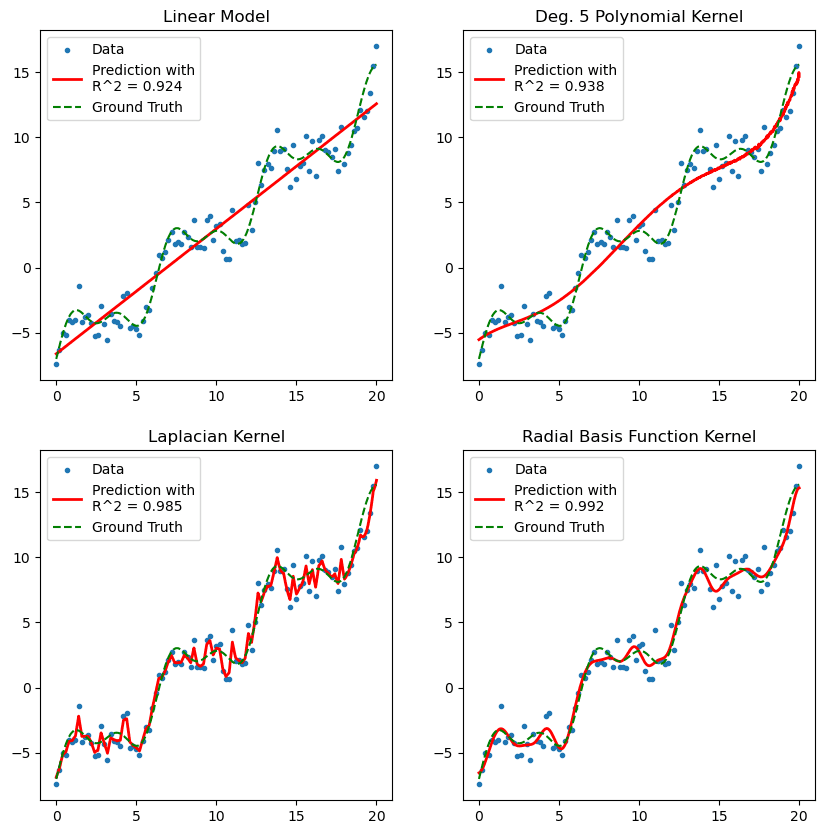
A few observations:
- The linear kernel under performs compared to our first linear model, as there is no built-in intercept.
- The degree-5 polynomial is constrained by dimension and therefore not expressive enough to capture the mixed periodicity in our ground truth data.
- The Laplacian kernel, whilst allowing an infinity dimensional feature representation, penalises higher order features.
- In practice, you shall most likely be turning to the Radial Basis Function (RBF), or Gaussian kernel, which here captures different scales of periodicity in the data trend.